The paper introduced in this presentation focuses on analyzing sperm motility heterogeneity via machine learning. With data from the Computer-Assisted Sperm Analysis (CASA) system as its core, it proposes leveraging supervised/unsupervised learning to explore motility subpopulations and innovates trajectory visualization methods to enhance identification accuracy. The paper breaks free from the dimensions of traditional analysis and retains comprehensive motility information; however, it is limited to 2D motility analysis and fails to establish a connection with subsequent fertilization and pregnancy outcomes. While providing a new pathway for reproductive biology research, it also identifies directions for future optimization.

Schematic diagram of the calculation process for motion parameters (VSL) of the CASA system
1. Research Background: Core Contradictions and Value
The Computer-Assisted Sperm Analysis (CASA) system is a core tool for analyzing sperm motility. It can generate raw 2D (x, y) motility coordinates (for trajectory construction) and 8–12 condensed motility parameters, laying the foundation for identifying sperm motility heterogeneity. Nevertheless, traditional research faces bottlenecks: univariate statistics cannot handle multi-dimensional data, easily masking heterogeneity; in addition, some early commercial CASA systems do not support the storage of raw coordinates, leading to the loss of motility information.
On one hand, traditional univariate statistical methods (e.g., only calculating the mean and dispersion of a single parameter) are incapable of processing the multi-dimensional data from CASA. They tend to overlook the correlations between parameters and even obscure the inherent motility heterogeneity of sperm (i.e., differences in sperm motility patterns within the same semen sample). On the other hand, early commercial CASA systems have limitations—some do not support the storage and export of raw coordinates, making it impossible for researchers to reconstruct trajectories afterward. As a result, researchers can only rely on condensed parameter analysis, which causes the loss of motility information.
Sperm motility heterogeneity is crucial for assisted reproductive technologies (both in human medicine and livestock breeding). Sperm from different motility subpopulations exhibit variations in their responses to environmental substances (such as bicarbonate and toxins) and their ability to bind to oocytes. Accurately analyzing this heterogeneity is key to optimizing semen dose production and improving reproductive efficiency.

Schematic diagram of sperm trajectory types and data augmentation representation
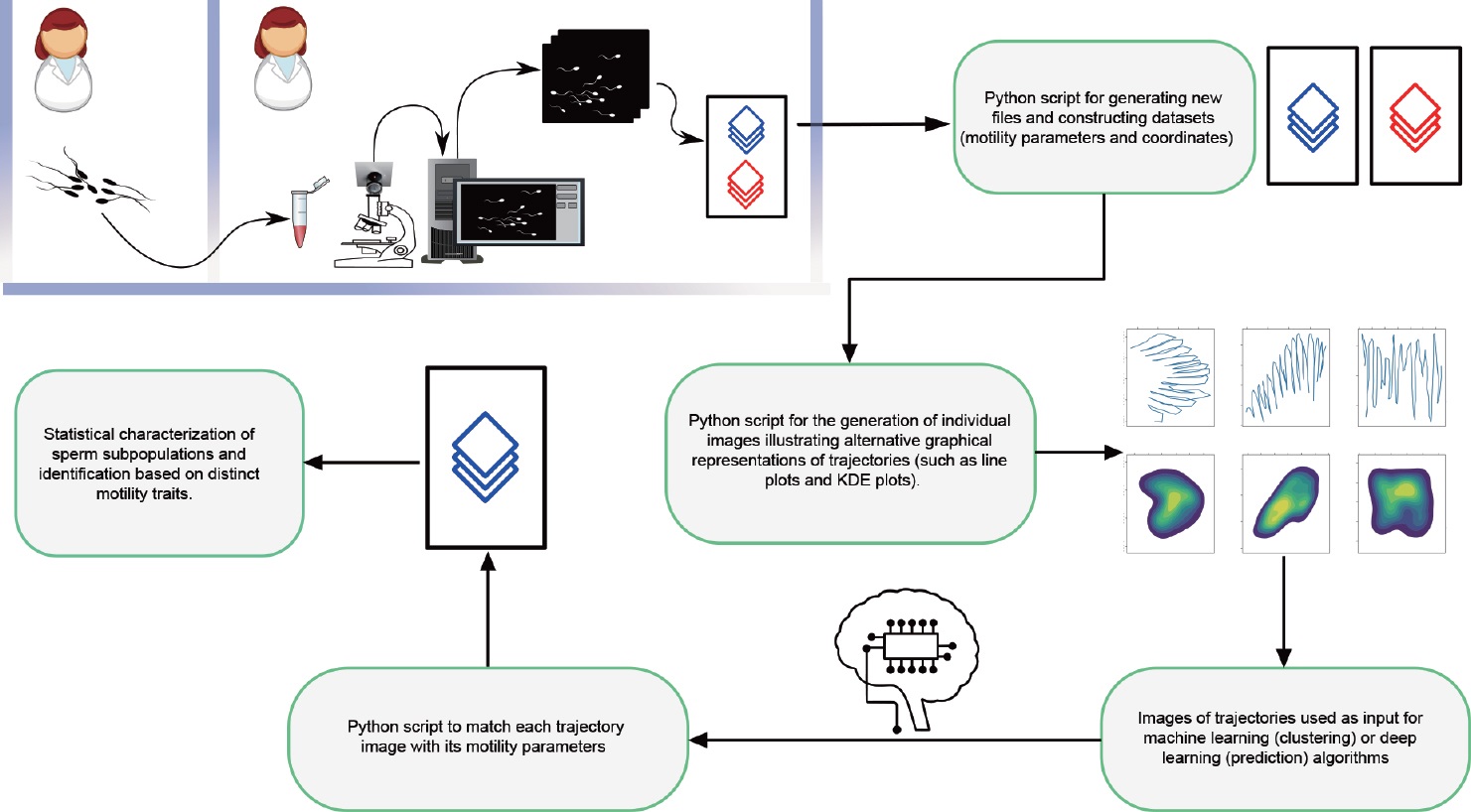
2. Technologies and Methods: Core Approaches
To obtain reliable sperm motility data, the study established a data collection system with the CASA system at its core: it utilized computer vision technology to achieve sperm detection and track (x, y) coordinates across consecutive frames. Combined with “pixel-micron” calibration and observation time, the system automatically calculated motility parameters (including velocity, linearity, and beat frequency) and generated trajectories. To address the high cost of commercial CASA systems and the inaccessibility of their raw coordinates, the research team developed open-source tools (e.g., CASA software, OpenCASA) based on the ImageJ platform. These tools support the simultaneous storage of coordinates and parameters, and are compatible with sperm analysis across multiple species (such as fish, horses, pigs, and humans), providing low-cost and comprehensive data support for subsequent research.
In the data processing phase, traditional statistical methods were first employed for exploration: Principal Component Analysis (PCA) was used to reduce the dimensionality of condensed motility parameters, followed by hierarchical clustering algorithms (either agglomerative or divisive) to classify motility subpopulations. Finally, descriptive and inferential statistics were combined to analyze the effects of treatments (e.g., pH adjustment, hormone application) on these subpopulations. However, this method relies on condensed parameters, making trajectory reconstruction impossible and resulting in information loss.
To overcome this limitation, the study introduced machine learning to optimize the analysis workflow:
- Supervised Learning: Using manually labeled sperm motility types (e.g., hyperactivated, motile) as training data, a Support Vector Machine (SVM) algorithm was adopted to automatically classify motility patterns based on CASA condensed parameters. Its effectiveness has been verified across multiple species, with the only drawback being the time-consuming nature of manual labeling.
- Unsupervised Learning: No labeling is required. Through a Python workflow, raw CASA coordinates are reconstructed into trajectory images, which are then input into clustering algorithms to automatically explore potential motility subpopulations.
Meanwhile, the study performed data augmentation via image transformations (e.g., random flipping, rotation) to improve model robustness. It also innovatively used Kernel Density Estimation (KDE) plots and heatmaps instead of traditional line graphs to represent trajectories, making fuller use of spatial information and particularly enhancing the identification accuracy of hyperactivated sperm.

A schematic diagram of the entire “data-model” process for trajectory image clustering
3. Research Results: Key Findings
Commercial CASA systems have obvious limitations: some do not open up raw coordinates, making it impossible to identify all motility subpopulations using only condensed parameters; others only provide parameter range values rather than individual sperm data, further reducing the accuracy of subpopulation identification. In contrast, open-source CASA tools effectively address these issues—they not only support raw coordinate storage but also are compatible with multi-species analysis. This provides comprehensive data support for machine learning and breaks through data bottlenecks.
Machine learning demonstrated remarkable application effectiveness: unsupervised learning can stably explore motility subpopulations and also capture the effects of environmental interventions (e.g., 5-hydroxytryptamine 2A receptor inhibition, tryptophan addition) on trajectories; supervised learning (SVM) achieves high accuracy in classifying human sperm motility patterns. When combined with enhanced trajectory representation methods such as KDE plots, the sensitivity of hyperactivated sperm identification is significantly improved, making up for the shortcomings of traditional threshold methods.
Bivariate analysis showed that when sperm are exposed to toxins (e.g., mercury chloride) or regulators (e.g., bicarbonate), a specific subpopulation with high VAP (Average Path Velocity) and high LIN (Linearity) emerges, confirming that motility heterogeneity is associated with environmental responses. The study also speculates that this heterogeneity may originate from the uneven distribution of molecules during spermatogenesis and epididymal maturation, and directly affects the efficiency of assisted reproduction—pointing out directions for subsequent mechanistic research.

Schematic diagram comparing traditional trajectory representation with data-augmented representation (KDE diagram)
4. Research Innovations: Core Breakthroughs
The study achieved multi-dimensional innovations in technical methods: first, it expanded the dimensions of analysis. Breaking free from the traditional reliance on condensed parameters, it used sperm trajectory images as input for machine learning for the first time. This retains comprehensive information such as trajectory morphology and spatial distribution, making the results more consistent with the actual state of sperm motility and avoiding information loss caused by parameter condensation.
Second, it innovated trajectory visualization methods. Traditional line graphs struggle to clearly present spatial patterns, while enhanced representation methods such as KDE plots and heatmaps can fully utilize 2D (x, y) spatial information, solving the problem of ambiguous information in line graphs. They perform particularly well in the identification of hyperactivated sperm, enabling accurate capture of this specific subpopulation.
Furthermore, it realized deep integration of open-source tools and AI: through open-source CASA tools, the data monopoly of commercial software is broken, reducing the cost of data acquisition; combined with Python, an integrated workflow of “data acquisition – trajectory reconstruction – AI analysis” is built, improving research reproducibility and lowering the threshold for studying sperm heterogeneity across multiple species.
5. Research Limitations and Future Outlook
The study still has limitations: in terms of data acquisition, some commercial CASA systems do not open up raw coordinates, restricting multi-center data sharing; in terms of machine learning, models rely on high-quality data, but the scale of existing labeled datasets is limited, and results from unsupervised learning require more biological validation; motility analysis is only limited to 2D, and 3D detection technologies (e.g., 3D imaging of sperm flagella) are expensive and less accessible, making widespread adoption difficult.
Future directions are clear: optimize open-source CASA tools to promote data standardization and cross-platform sharing; expand labeled datasets to improve the cross-species generalization ability of models; explore the clinical application of enhanced trajectory representation methods and develop an “automatic high-quality sperm screening” system; promote the cost reduction of 3D technology and integrate it into existing systems to achieve comprehensive analysis of sperm motility, providing precise support for reproductive research and fertility assessment.
Reference
Aragón-Martínez, Andrés. Unraveling sperm kinematic heterogeneity with machine learning. Asian Journal of Andrology ():10.4103/aja202544, August 12, 2025. | DOI: 10.4103/aja202544