In the clinical practice of assisted reproductive technology (ART), embryo quality assessment is directly linked to embryo transfer decisions and live birth outcomes. Although existing assessment protocols have been widely applied in clinical settings, the overall live birth rate has long remained at approximately 30%. This indicates that morphological observation at limited time points may be insufficient to fully reflect the developmental potential of embryos. With the advancement of microscopic imaging technology, continuous bright-field microscopic images can be acquired during embryo culture, providing the conditions for observing dynamic changes throughout embryonic development. However, it remains a practical challenge to extract stable and reproducible quantitative information from these images without introducing additional interventions. The paper introduced herein explores how to leverage dynamic information during embryonic development to provide a new quantitative approach for evaluating embryo live birth potential, without altering conventional culture and bright-field imaging protocols.
Kanazawa T, Yao T, Takeshita S, et al. Nuclear segmentation in four-dimensional label-free microscopy images for predicting live birth potential of mouse embryos. Comput Biol Med. 2025;198(Pt A):111179. doi:10.1016/j.compbiomed.2025.111179.
Research Methods
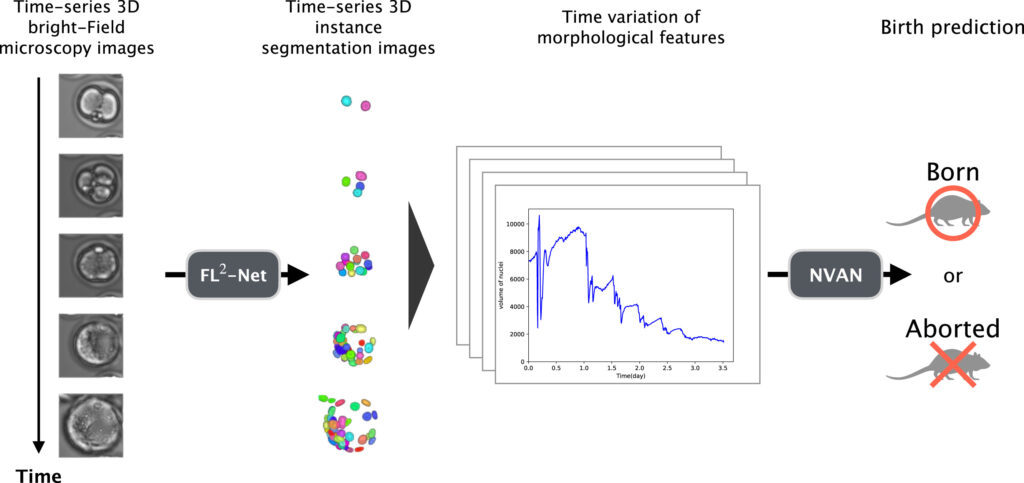
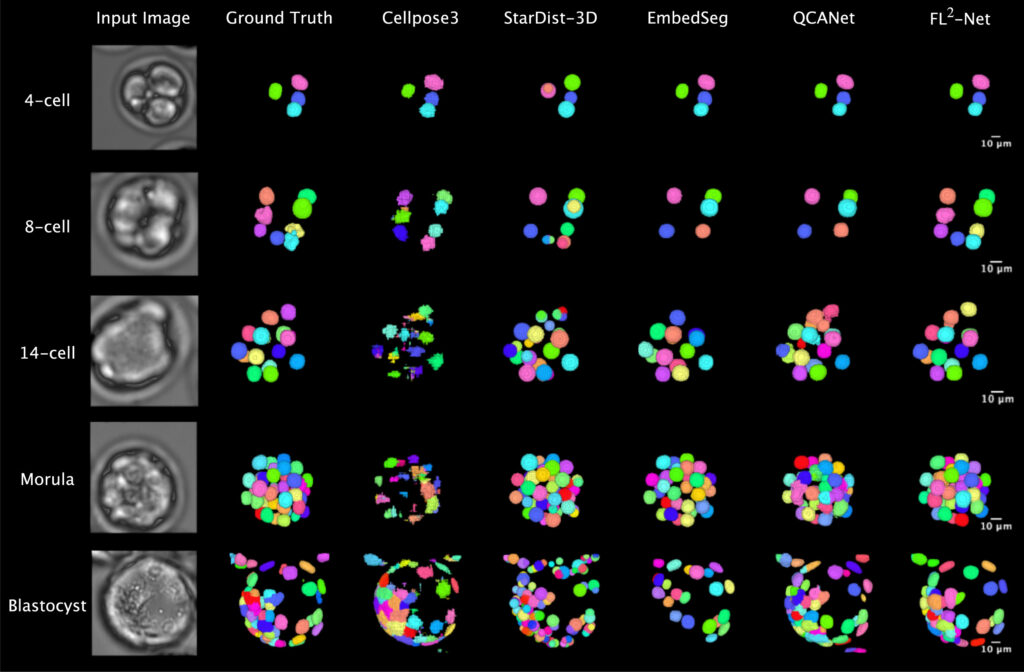
In response to the imaging characteristics of bright-field microscopic images, such as low nuclear contrast and high cellular density at the blastocyst stage, the research team designed a 4D analysis pipeline for mouse embryos to observe the continuous developmental process from fertilization to blastocyst formation. The term 4D here refers to the integration of temporal developmental dynamics with three-dimensional spatial structure. In the experimental design, method construction and outcome validation were conducted separately: 84 mouse embryos were enrolled to establish the nuclear recognition method, and an additional 147 embryos were used for independent validation of live birth outcomes, avoiding simultaneous modeling and evaluation on the same dataset.
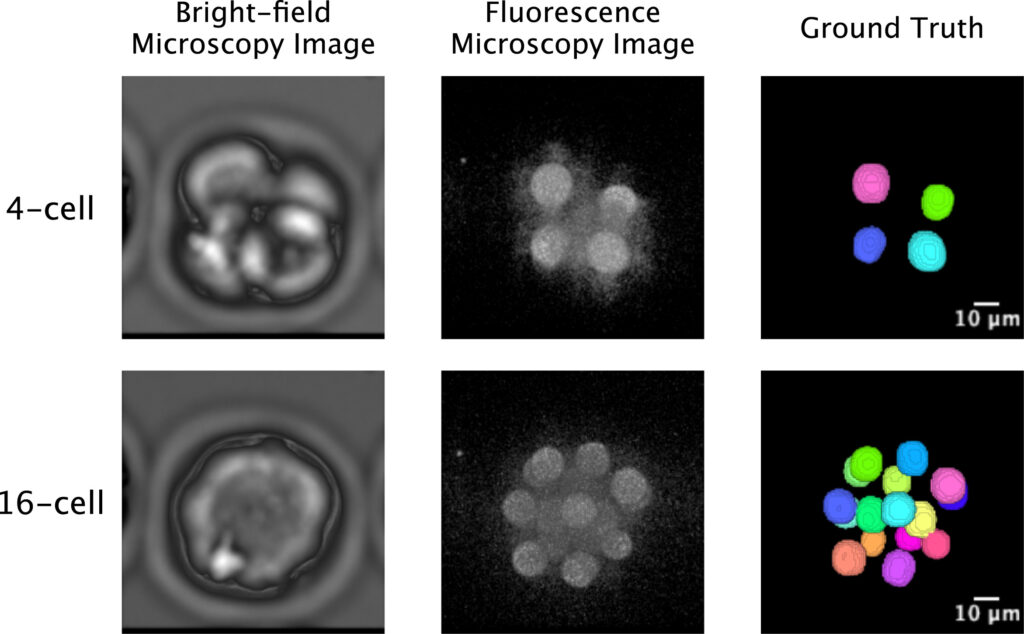
Since clear nuclear reference information cannot be directly obtained from bright-field images alone, fluorescent imaging was introduced as a control during the method construction phase to establish a reliable annotation basis. Manual correction of nuclear segmentation results under fluorescent conditions provided relatively accurate reference standards for the corresponding bright-field images. It is important to emphasize that fluorescent labeling was only used for method establishment and validation, and was not involved in subsequent analysis and prediction based on bright-field images.
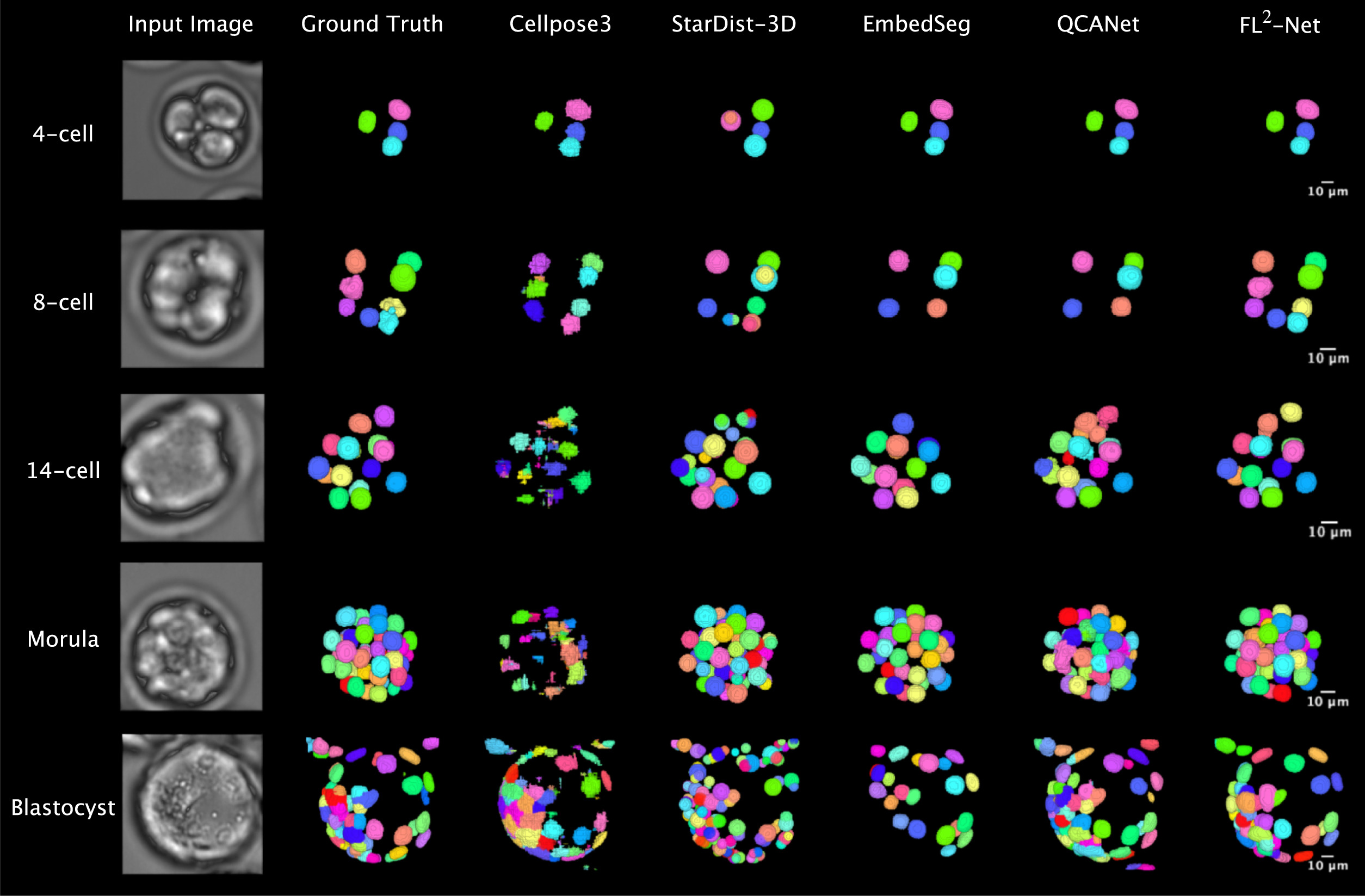
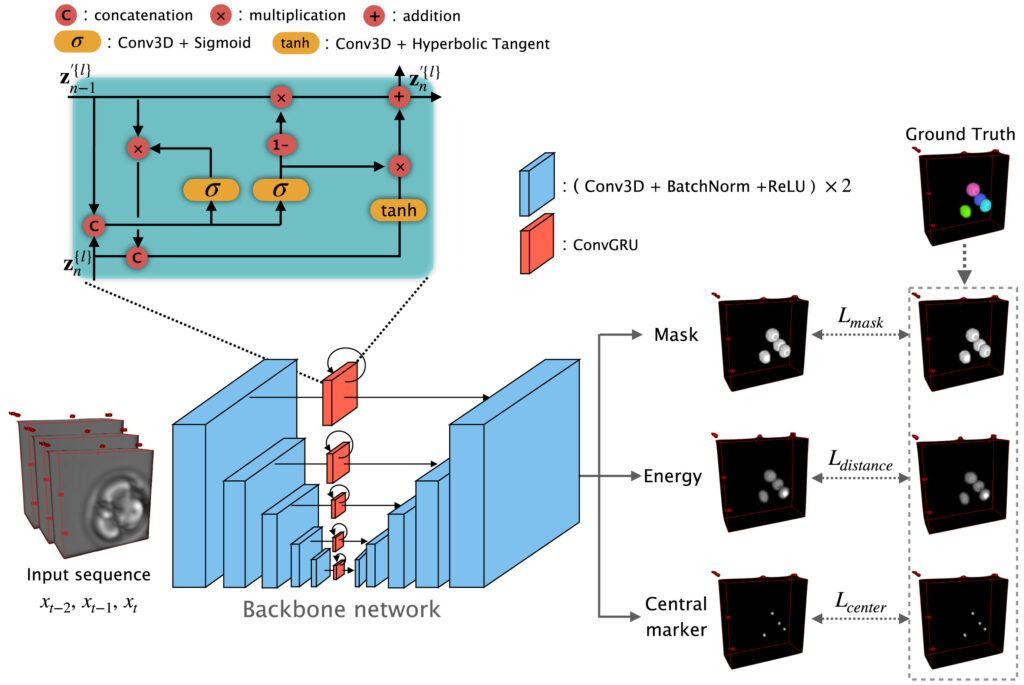
On this basis, the research proposed the FL²-Net model for analyzing continuous microscopic images during embryonic development. While processing images at the current time point, the model incorporates information from adjacent temporal frames to capitalize on the continuity of embryonic development and enhance the stability of nuclear recognition under low-contrast conditions. For the high cellular aggregation observed at the blastocyst stage, the model can distinguish closely adjacent cell nuclei and obtain complete nuclear contour information. Furthermore, 11 categories of morphological indicators, including nuclear count, volume, surface area and their temporal changes, were extracted for live birth potential analysis.
Research Results
In the evaluation of nuclear segmentation performance, this method exhibited stable recognition efficacy across different embryonic developmental stages and outperformed several commonly used 3D segmentation methods on multiple evaluation metrics. As embryos developed to the blastocyst stage at approximately 2.5 days post-fertilization, with a marked increase in cell number and more compact spatial structure, the segmentation accuracy of several control methods decreased, whereas this method maintained relatively consistent segmentation performance at this stage. Further comparative experiments showed that after incorporating continuous temporal information, the model had little difference from time-agnostic models in the early developmental stages, but its segmentation stability and consistency were more pronounced under the condition of high cellular density at the blastocyst stage.
For the live birth potential prediction task, a predictive model was constructed based on morphological features such as nuclear count, volume, surface area and their temporal changes, achieving an approximate prediction accuracy of 81.6% on an independent test dataset. Feature analysis results revealed that the variability of nuclear volume had a high weight in multiple predictions, and its influence was more prominent in the early stages of embryonic development, suggesting that the stability of nuclear morphological changes during cell division may be associated with subsequent developmental outcomes.
Innovations and Limitations
This study proposed a label-free 4D embryonic analysis pipeline based on conventional bright-field microscopic imaging, enabling the continuous quantification of nuclear changes during embryonic development without the introduction of exogenous dyes. In contrast to static assessment methods primarily based on single time points, this method focuses more on the developmental process itself, and by integrating time-series information, it maintains stable analytical performance even with high cellular density at the blastocyst stage. Additionally, the research correlates the key features focused on by the model with specific morphological indicators, making it easier to establish a connection between analytical results and the embryonic development process.
It should be viewed objectively that this study was conducted based on mouse embryo data. Although the overall process of early embryonic development in mice and humans shares certain similarities, there are still differences in developmental rhythm, oocyte size and nuclear morphology. Further validation is required before the relevant method can be applied to human embryo assessment. Moreover, this method has a certain dependence on imaging quality and temporal resolution, and its stability under different clinical equipment and imaging conditions remains to be evaluated. Current analyses mainly focus on nuclear-level features and have not incorporated other information such as cytoplasmic structure and zona pellucida, leaving room for further expansion in subsequent research.
Significance and Future Outlook
Overall, this study demonstrates a technical approach for the continuous quantitative analysis of embryonic development under conventional bright-field microscopic imaging conditions. By focusing on the relatively stable structure of the cell nucleus and integrating temporal information, this method is expected to serve as an auxiliary tool for traditional morphological assessment, helping to reduce inter-observer variability in judgment.
With the gradual accumulation of more human embryo data under clinically relevant conditions and the integration of more subcellular structural information, such 4D dynamic analysis-based methods are expected to be further refined, providing more stable and interpretable support for embryo selection in assisted reproductive technology.