The paper introduced herein focuses on a critical evaluation bottleneck in the field of in vitro fertilization (IVF) embryo selection: in clinical practice, untransferred embryos cannot be observed for the outcome of “whether they would result in a live birth if transferred” because they were not selected. To address this challenge, the study innovatively incorporates multiple imputation, for the first time integrating untransferred embryos into a comprehensive evaluation system. Additionally, the research constructs two complementary AUC metrics to accurately measure ranking performance: the population-level AUC, which spans across different patients and treatment cycles, and the treatment-level AUC, which focuses on embryo cohorts of the same patient within a single cycle. These provide new insights for evaluating embryo selection algorithms.
The Dilemma in IVF Embryo Selection: Exploring Accurate Evaluation
In in vitro fertilization (IVF), time to live birth (TTLB), defined as the number of embryo transfers required to achieve a live birth, is a core indicator of treatment efficacy, and the accuracy of embryo selection directly impacts this metric. Traditional methods relying on Gardner morphological grading are highly subjective with limited ranking precision. Emerging AI algorithms (such as iDAScore) show potential, but evaluating their clinical utility faces a critical obstacle: the live birth outcomes of untransferred embryos are inherently missing, making it impossible to fully validate their ranking value in real clinical scenarios. This study aims to address outcome missingness through innovative methods, objectively compare the clinical utility of iDAScore and Gardner grading, and provide a basis for optimizing embryo selection algorithms.
The Challenge of Missing Outcomes: A Breakthrough via Multiple Imputation
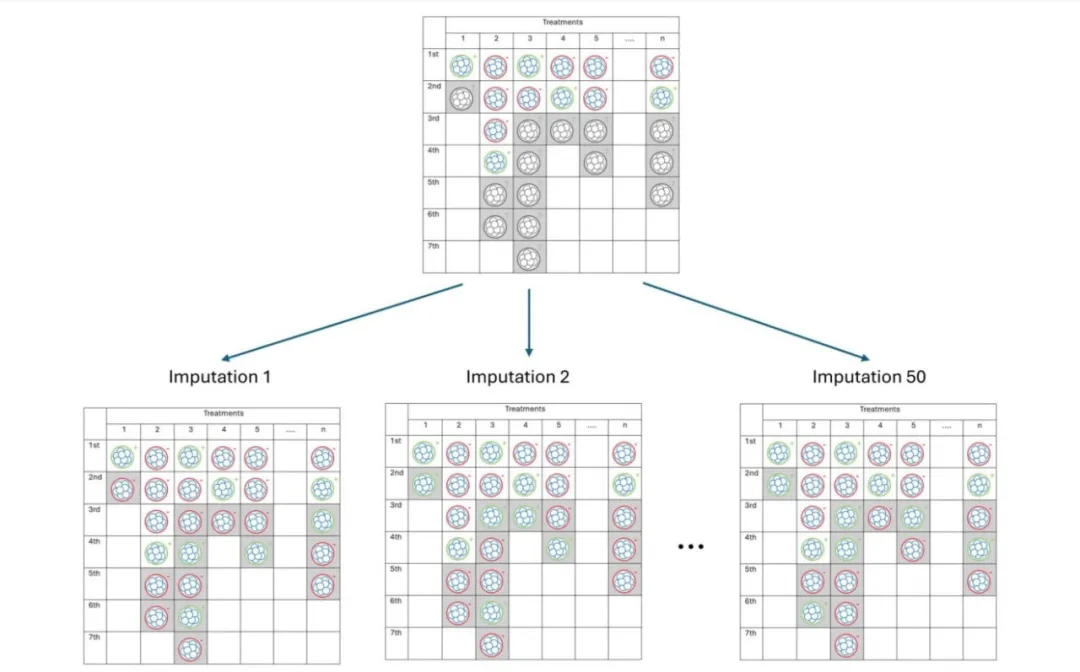
The core challenge in evaluation lies in missing live birth outcomes of untransferred embryos—since these embryos are not selected in clinical practice, information on “whether they would result in a live birth if transferred” is unknown, making it impossible to reconstruct complete clinical selection scenarios. To tackle this, the study employs multiple imputation by chained equations (MICE): based on variables closely related to embryo viability (patient age, oocyte origin, inner cell mass (ICM) and trophectoderm (TE) quality grades, previous transfer outcomes, etc.), a random forest model is used to simulate potential live birth outcomes of untransferred embryos (0 = non-live birth, 1 = live birth).
Robustness in the imputation process is achieved by generating 50 independent datasets: each imputation randomly samples results from the model to reflect outcome uncertainty, while 50 datasets address statistical biases caused by high missing rates (untransferred embryos account for 57.7%), ensuring the reliability of subsequent analyses (e.g., TTLB and AUC calculations). Imputed outcomes must align with clinical logic—for example, imputed live birth rates are higher for younger patients and higher-grade embryos, and the imputed live birth rate of untransferred embryos (33.2%) is lower than that of transferred embryos (42.8%), consistent with the clinical practice of “prioritizing transfer of high-quality embryos.”

Measuring Ranking Performance: Distinctions and Calculations of Two AUCs
The study evaluates the ranking performance of algorithms using two types of area under the receiver operating characteristic curve (AUC), which differ significantly due to their distinct evaluation perspectives.
Population-level AUC focuses on overall ranking performance across treatment cycles, i.e., the algorithm’s accuracy in distinguishing live births from non-live births among “all embryos (from different patients).” Its calculation is based on the scores/grades of all embryos (transferred + imputed untransferred) and their corresponding outcomes: using scores/grades as thresholds, sensitivity (proportion of correctly identified live birth embryos) and specificity (proportion of correctly identified non-live birth embryos) are calculated at different thresholds to construct an ROC curve, and the area under the curve is then computed. The Obuchowski method is used to correct for clustering correlation among embryos from the same patient, avoiding result biases caused by inter-embryo associations.

Treatment-level AUC, on the other hand, targets ranking performance within a single treatment cycle, i.e., the algorithm’s accuracy in distinguishing live births from non-live births among “an embryo cohort of the same patient” (closer to real clinical selection scenarios). Its calculation is restricted to “non-trivial cycles” (with at least 1 live birth and 1 non-live birth embryo): within a single cycle, all live birth embryos are paired with non-live birth embryos, and the proportion of pairs where “the live birth embryo has a score/grade ≥ the non-live birth embryo” (i.e., the correct ranking proportion) is calculated, which serves as the AUC for that cycle. The overall treatment-level AUC is obtained by averaging AUCs across all cycles and pooling results from 50 imputed datasets using Rubin’s rules.
Key Result Comparisons: Significant Advantages of the AI Algorithm
Comparisons of iDAScore and Gardner grading performance show: in terms of TTLB, the average TTLB for iDAScore ranking is 1.68 transfers, 6.1% shorter than that for Gardner grading (1.78 transfers), with the advantage becoming more pronounced as the number of embryos increases (e.g., a 10.7% reduction in cycles with 7 embryos). In terms of ranking performance, iDAScore (0.633) slightly outperforms Gardner grading (0.619) in population-level AUC; in treatment-level AUC, iDAScore (0.672) significantly outperforms Gardner grading (0.631). Notably, treatment-level AUC for both methods is higher than population-level AUC, indicating that traditional cross-cycle evaluation underestimates actual ranking performance. Validation of imputed data shows that the associations between simulated outcomes of untransferred embryos and patient age/embryo quality align with clinical patterns, confirming the rationality of imputation.

Innovation and Limitations: Real-World Constraints Amid Breakthroughs
The innovations of this study lie in: for the first time, achieving comprehensive evaluation of all embryos (including untransferred ones) through multiple imputation, addressing the core challenge of missing outcomes; distinguishing the two AUCs to reveal limitations of traditional evaluation, providing a more clinically relevant measure of algorithm performance; and quantitatively validating the advantages of AI algorithms in reducing TTLB and improving ranking precision.
The study also has limitations: imputed outcomes depend on the quality of input variable assessment, potentially introducing model bias; results are based on single-center data (73% from donated oocytes), limiting generalizability; a maximum of 7 transfers are included, failing to cover scenarios with more transfers; untransferred embryos are imputed based on iDAScore ranking, which may slightly overestimate its performance.
Clinical Value and Prospects
Clinically, the advantage of iDAScore in reducing TTLB can alleviate patient treatment burdens, and its higher treatment-level AUC facilitates precise selection of high-potential embryos, optimizing clinical decision-making. Methodologically, multiple imputation provides a paradigm for handling outcome missingness in assisted reproduction, which can be extended to other algorithm evaluations. Future research should validate generalizability through multi-center studies, confirm the value of AI algorithms via prospective trials, and explore optimization of “AI + manual assessment” combined strategies.
reference
Bori L, Johansen MN, Berntsen J, et al. Predicting time to live birth with deep learning embryo ranking: a novel multiple imputation approach. Hum Reprod. 2025 Jun 13. doi:10.1093/humrep/deaf102